 |
 |
- Search
| J Audiol Otol > Volume 28(2); 2024 > Article |
|
Abstract
Cochlear implantation is currently the treatment of choice for children with severe-to-profound sensorineural hearing impairment (SNHI). However, the outcomes with cochlear implant (CI) vary significantly among recipients. Genetic diagnosis offers direct clues regarding the pathogenesis of SNHI, which facilitates the development of personalized medicine for potential candidates for CI. In this article, I present a comprehensive overview of the usefulness of genetic information in clinical decision-making for CI. Genetically confirmed diagnosis enables clinicians to: 1) monitor the evolution of SNHI and determine the optimal surgical timing, 2) predict the potential benefits of CI in patients with identified genetic etiology, and 3) select CI devices/electrodes tailored to patients with specific genetic mutations.
Sensorineural hearing impairment (SNHI) stands as the leading sensory deficit in children, affecting approximately 1.9 per 1,000 live births [1], with an additional 1 per 100 school-aged children presenting with late-onset SNHI [2]. Genetic factors contribute to more than 50% of SNHI cases, with more than 200 genes identified to date [3]. For children with mild or moderate SNHI, hearing aid fitting is the primary intervention. However, in cases of profound SNHI, cochlear implantation becomes mandatory.
Advances in genomic medicine, particularly next-generation sequencing (NGS), have significantly facilitated the integration of genetic examination into clinical practice. NGS-based genomic sequencing has gradually replaced traditional genetic testing and has become the main diagnostic tool for pediatric SNHI [4-7]. In combination with imaging and virological studies, NGS allows etiologic identification in approximately 80% of pediatric cochlear implant (CI) candidates [8-10]. This includes approximately 50% of cases with non-syndromic SNHI, approximately 10% with syndromic SNHI (e.g., Waardenburg syndrome), approximately 10% with acquired SNHI (e.g., congenital cytomegalovirus infection), and approximately 10% with inner ear malformations (e.g., cochlear nerve deficiency) (Fig. 1) [8,9].
Management of pediatric SNHI involves sequential clinical decisions that take into account the severity and progression of hearing loss. The potential for progressive hearing loss prompts consideration of the need for cochlear implantation and the optimal timing of such intervention. If cochlear implantation is subsequently deemed necessary, an assessment of the potential benefits to the patient becomes paramount. The final decision to proceed with surgery involves careful selection of the most appropriate device or electrode for the individual patient. Genetic information can provide critical insight into these decision-making processes.
Genetic diagnosis plays a pivotal role in unraveling the natural progression of SNHI and determining the ideal timing for cochlear implantation, closely aligning with the clinical course. Different pathogenic variants are associated with different patterns of SNHI progression; for example, GJB2 mutations are associated with stable or slow progression [11-13], whereas SLC26A4 mutations are associated with fluctuating SNHI [14-16].
The close relationship between genetic diagnosis and clinical progression allows prediction of the timing of CI surgery. In a previous study, we formulated a prediction model for GJB2-related SNHI [17]. Our results showed the severity of hearing loss in individuals with pathogenic GJB2 variants correlated with their genotypes and baseline hearing levels. On average, the rate of hearing loss deterioration was approximately 0.55 dB per year, as shown in Fig. 2 [17]. This observation was corroborated in a recent study that reported a rate of deterioration of approximately 0.3 dB per year in patients with pathogenic GJB2 variants [18].
This predictive model serves as a tool to estimate when a patientŌĆÖs hearing may deteriorate to the point where cochlear implantation may be required. For example, consider a 3-year-old child homozygous for the GJB2 c.235delC variant (Fig. 2B, case #3) with an initial hearing level of 85 dB HL. Given that the eligibility criterion for cochlear implantation is 90 dB HL, the model predicts that approximately 9 years later, the childŌĆÖs hearing loss is likely to reach the 90 dB HL threshold, making him or her eligible for insurance coverage of a CI.
In addition to predicting disease progression, genetic diagnosis plays a critical role in guiding treatment decisions, particularly those related to cochlear implantation. Our extensive genetic studies involving more than 600 CI patients in Taiwan have revealed clear associations between genetic mutations and CI outcomes (Fig. 3) [8,19-21]. Pathogenic variants in most genes, such as GJB2, SLC26A4, and MYO15A, are associated with favorable CI outcomes, likely due to confined pathology within the inner ear [8,19-21]. These findings are consistent with other series [10,22] and have important clinical implications. Identification of pathogenic variants in these genes serves as a reliable predictor of CI outcomes, expediting the decision-making process for otologists, audiologists, geneticists, and patients.
Conversely, pathogenic variants in genes such as PJVK and PCDH15 are associated with less favorable CI outcomes [23], likely due to involvement of spiral ganglion neurons [24]. Notably, pediatric patients with pathogenic PJVK and PCDH15 variants presented with clinical features indistinguishable from those of other typical pediatric CI recipients, underscoring the need for genetic testing for accurate diagnosis [23].
Genetic diagnosis is particularly valuable in the management of auditory neuropathy spectrum disorder (ANSD), which constitutes approximately 10% of pediatric SNHI cases [25]. The heterogeneous pathologies of ANSD range from inner hair cells to the auditory cortex [26]. Our previous study highlighted that acquired factors such as prematurity and kernicterus are the predominant contributors to pediatric ANSD, while genetic factors account for approximately 1/5 of cases, with pathogenic OTOF variants being the primary genetic cause [27].
Depending on the location of the lesion, ANSD is classified as pre-synaptic or post-synaptic. In general, pre-synaptic ANSD is associated with positive CI outcomes, whereas post-synaptic ANSD is associated with less favorable CI outcomes [26]. The exact location of the pathology is critical. For instance, OTOF variants primarily affect the synapse, leaving the neurons intact for CI function. Consequently, patients with pathogenic OTOF variants exhibit robust electrically evoked compound action potential responses during surgery and excellent hearing and speech performance after surgery [28]. Therefore, early cochlear implantation is recommended for ANSD patients with pathogenic OTOF variants.
In a recent study at the National Taiwan University Hospital, we analyzed CI outcomes in 36 ANSD patients, including 18 with pathogenic OTOF variants, 2 with rare pathogenic variants in the OPA1 and WFS1 genes, 10 with cochlear nerve deficiency (CND) and 6 without a definite diagnosis (Fig. 4) [29]. Aided behavioral thresholds showed improvement in all patients after surgery, although they were higher in the CND group. In particular, patients with OTOF, WFS1, and OPA1 variants showed favorable CI outcomes, as evidenced by good categorical auditory performance and speech intelligibility rating scores. In contrast, patients with CND had suboptimal outcomes, highlighting the importance of identifying ANSD etiologies prior to surgery for prognostication and counseling.
In addition to ANSD, genetic information can guide clinical decision-making for patients with unilateral or asymmetric SNHI, a condition for which cochlear implantation has emerged as a viable treatment option [30,31]. However, similar to bilateral SNHI, the etiologies of unilateral or asymmetric SNHI are remarkably diverse. In our recent studies, we analyzed the genetic basis in patients with unilateral or asymmetric SNHI. Our results showed that genetic causes could be identified in about 30% of patients with asymmetric SNHI (Fig. 5) [32]. Notably, genetic causes were also identified in approximately 10% of patients with unilateral SNHI [33], which was traditionally thought to be less likely to have a genetic origin. Specifically, pathogenic variants associated with Waardenburg syndrome were found to be a significant contributor to unilateral severe-to-profound SNHI [33]. This finding has significant clinical implications by suggesting a favorable candidacy for cochlear implantation, as the pathology of Waardenburg syndrome primarily affects the stria vascularis while leaving the cochlear nerve intact [34].
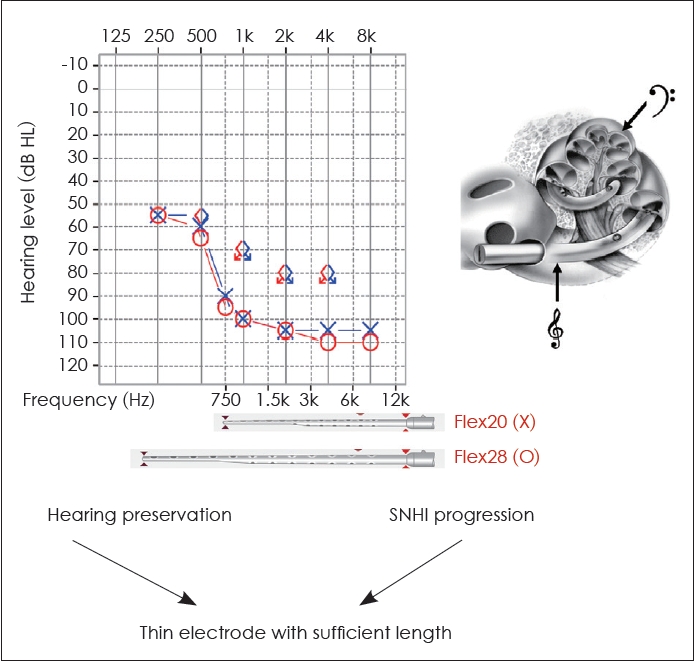
Genetic diagnosis plays a critical role in enabling clinicians to make informed decisions about electrode selection for cochlear implantation. Preservation of low and mid-frequency hearing has been observed in the early stages of certain genetic hearing impairments, such as pathogenic variants in the genes MYO15A [35,36], CDH23 [37], or TMPRSS3 [38]. However, a significant proportion of this residual hearing tends to gradually deteriorate over time [35-38]. Therefore, for individuals with MYO15A, CDH23, or TMPRSS3 variants seeking cochlear implantation, electrode selection must consider both the preservation of residual hearing and the progression of SNHI, with a thin electrode of sufficient length being the preferred choice. For example, when using MED-EL electrodes, the Flex20 electrode should be avoided. While the Flex20 may be beneficial for preserving hearing, it cannot cover the low-frequency range if residual low-frequency hearing is lost in the future. In such cases, the Flex28 electrode would be a more appropriate choice for the patient (Fig. 6). Thus, incorporating genetic findings into the selection of CI devices/electrodes allows for a more personalized, patient-specific auditory treatment.
In summary, our research over the past two decades underscores the critical role of genetic information in guiding clinical decisions related to cochlear implantation. The impact of genetic knowledge extends to multiple aspects of patient management, allowing the tracking of SNHI evolution to determine optimal surgical timing. In addition, the predictive power of genetic information is proving valuable in assessing the potential benefit of CI for diverse patient populations, including those with ANSD or unilateral SNHI. Finally, our findings highlight the importance of tailoring CI electrode selection based on specific genetic mutations, providing a more personalized approach to improving the efficacy of cochlear implant interventions.
Acknowledgments
In memory of Prof. Seung-Ha Oh, who has been a mentor and good friend of the author for the past two decades.
Fig.┬Ā1.
Etiologies of sensorineural hearing impairment (SNHI) in cochlear implant candidates in Taiwan. Next-generation sequencing, in combination with imaging and virological studies, facilitates the identification of etiologies in approximately 80% of pediatric cochlear implant candidates. This includes approximately 50% of cases with non-syndromic SNHI (represented by the red area), approximately 10% with syndromic SNHI (represented by the blue area), approximately 10% with acquired SNHI (represented by the green area), and approximately 10% with inner ear malformations (represented by the orange area).

Fig.┬Ā2.
Prediction of timing of cochlear implantation using genetic information. A: Prediction model for GJB2-related hearing impairment using multivariate generalized estimating equation analyses. Hearing level is estimated as: [predicted hearing level (dB HL)] = 3.78 + 0.96├Ś [baseline hearing level (dB HL)] + 0.55├Ś[duration of follow-up (yr)] [17]. B: The estimated time for the patient to meet cochlear implantation eligibility criteria can be determined based on the patient's age, genotype, and baseline hearing level.
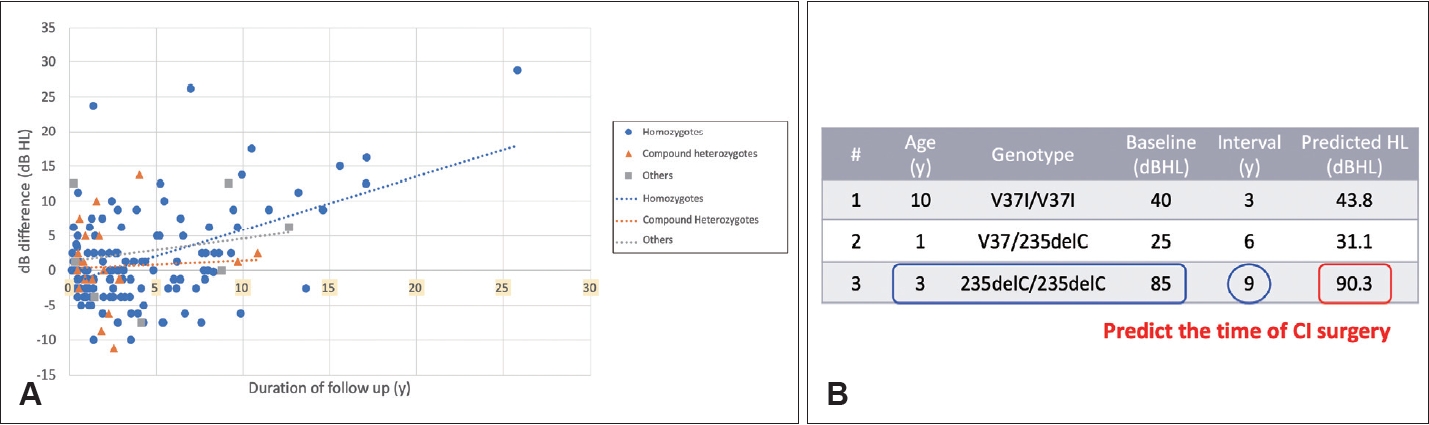
Fig.┬Ā3.
Relationship between genetic mutations and CI outcomes. Based on comprehensive genetic studies of more than 600 cochlear implant patients in Taiwan, our results indicate that pathogenic variants in several genes, including GJB2, SLC26A4, and MYO15A, are correlated with favorable CI outcomes, probably due to the confined pathology within the inner ear. Conversely, we observed that pathogenic variants in certain genes, such as PJVK and PCDH15, are associated with unfavorable CI outcomes, possibly due to the involvement of spiral ganglion neurons. CI, cochlear implant; NGS, next-generation sequencing; WES, whole exome sequencing; WGS, whole genome sequencing.
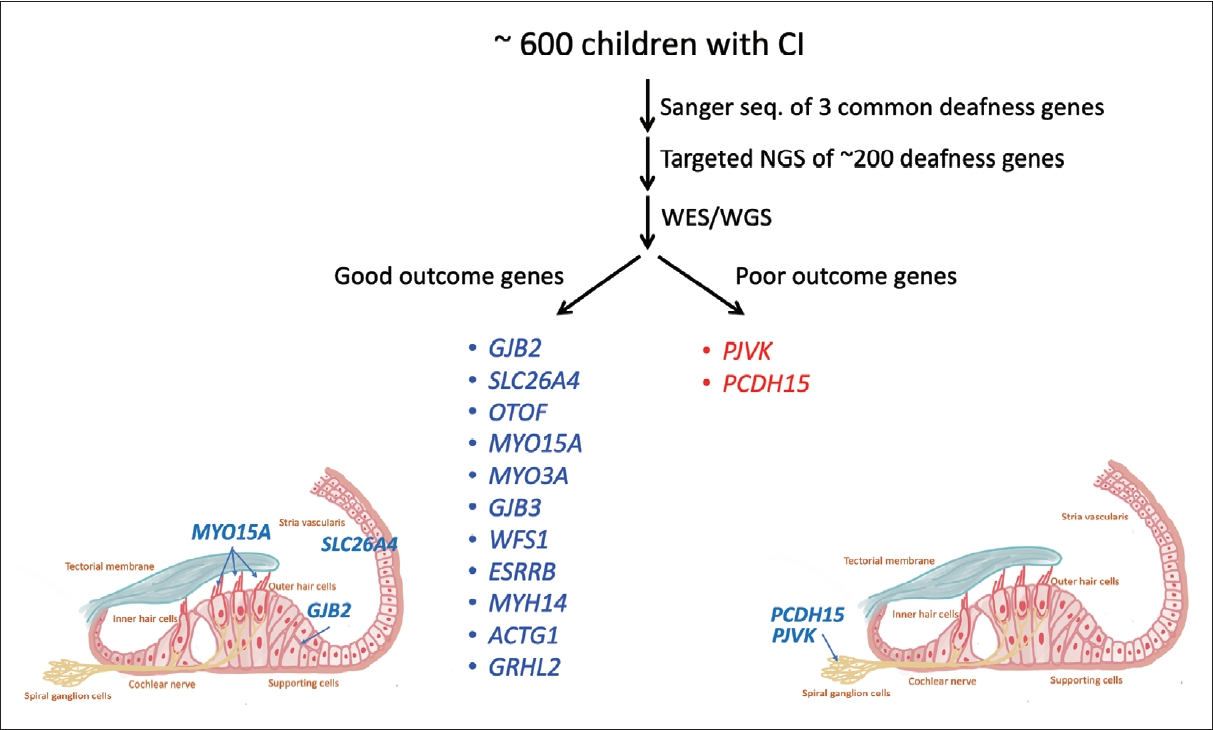
Fig.┬Ā4.
CI outcomes in patients with ANSD. Auditory and speech performance were analyzed in 36 ANSD patients who underwent cochlear implantation at the National Taiwan University Hospital, including 18 with OTOF mutations, 2 with rare mutations in OPA1 and WFS1 genes, 10 with CND, and 6 without definite diagnosis. Aided behavioral thresholds improved in all patients after surgery, but were higher in the CND group. Patients with OTOF, WFS1, and OPA1 had favorable CI outcomes with good CAP and SIR scores, whereas patients with CND had suboptimal CAP and SIR scores. ANSD, auditory neuropathy spectrum disorder; CAP, categorical auditory performance; CI, cochlear implant; CND, cochlear nerve deficiency; SIR, speech intelligibility rating.
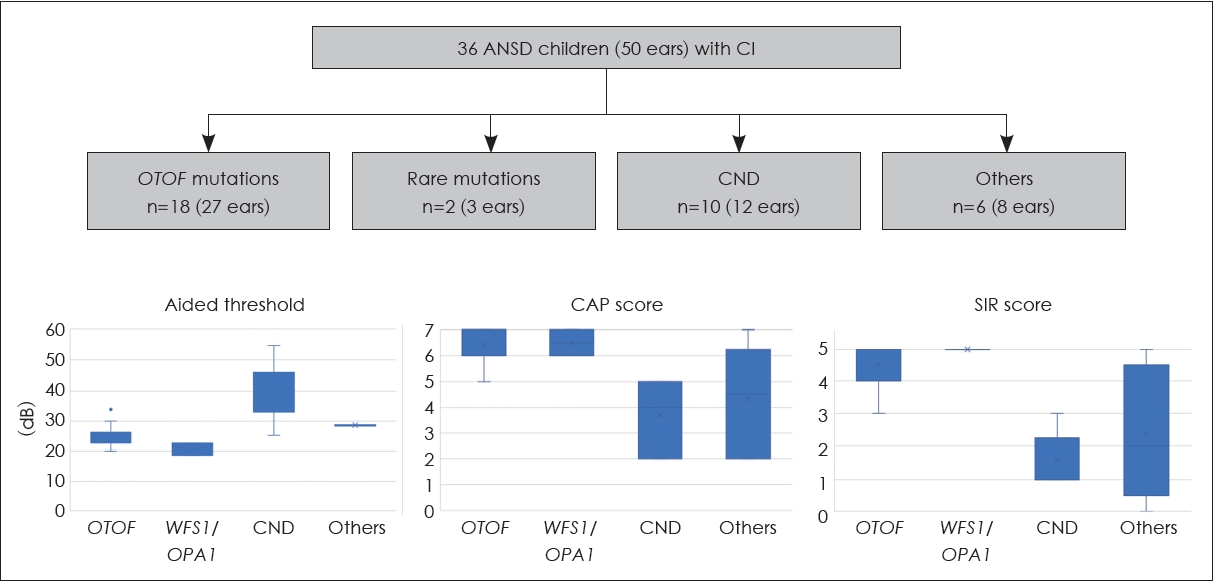
Fig.┬Ā5.
Genetic contribution to unilateral or asymmetric sensorineural hearing impairment. Using next-generation sequencing-based diagnostics, genetic causes can be identified in approximately 10% and 30% of patients with unilateral and asymmetric sensorineural hearing impairment, respectively. AHL, asymmetric hearing loss; UHL-RH, unilateral hearing loss with residual hearing; UHL-SO, unilateral hearing loss with scaled-out levels.
Fig.┬Ā6.
CI electrode selection in patients with MYO15A, CDH23, or TMPRSS3 mutations. In patients with pathogenic variants in MYO15A, CDH23, or TMPRSS3, electrode selection must carefully balance the preservation of residual hearing with the progression of sensorineural hearing loss. For optimal results, a thin electrode of sufficient length is recommended. Therefore, the Flex28 may be a more appropriate option than the Flex20 for these particular patients. SNHI, sensorineural hearing impairment; CI, cochlear implant.
REFERENCES
1. Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med 2006;354:2151ŌĆō64.


2. Wake M, Tobin S, Cone-Wesson B, Dahl HH, Gillam L, McCormick L, et al. Slight/mild sensorineural hearing loss in children. Pediatrics 2006;118:1842ŌĆō51.



3. Sheffield AM, Smith RJH. The epidemiology of deafness. Cold Spring Harb Perspect Med 2019;9:a033258



4. Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J 2nd, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A 2010;107:21104ŌĆō9.



5. Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol 2012;13:245



6. Wu CC, Lin YH, Lu YC, Chen PJ, Yang WS, Hsu CJ, et al. Application of massively parallel sequencing to genetic diagnosis in multiplex families with idiopathic sensorineural hearing impairment. PLoS One 2013;8:e57369.



7. Shearer AE, Smith RJ. Massively parallel sequencing for genetic diagnosis of hearing loss: the new standard of care. Otolaryngol Head Neck Surg 2015;153:175ŌĆō82.


8. Lee CY, Lin PH, Tsai CY, Chiang YT, Chiou HP, Chiang KY, et al. Comprehensive etiologic analyses in pediatric cochlear implantees and the clinical implications. Biomedicines 2022;10:1846



9. Miyagawa M, Nishio SY, Usami S. A comprehensive study on the etiology of patients receiving cochlear implantation with special emphasis on genetic epidemiology. Otol Neurotol 2016;37:e126. ŌĆō34.



10. Han JH, Kim SH, Moon IS, Joo SY, Kim JA, Gee HY, et al. Comprehensive prediction model, including genetic testing, for the outcomes of cochlear implantation. Ear Hear 2023;44:223ŌĆō31.


11. Chan DK, Schrijver I, Chang KW. Connexin-26-associated deafness: phenotypic variability and progression of hearing loss. Genet Med 2010;12:174ŌĆō81.


12. Wu CC, Tsai CH, Hung CC, Lin YH, Lin YH, Huang FL, et al. Newborn genetic screening for hearing impairment: a population-based longitudinal study. Genet Med 2017;19:6ŌĆō12.



13. Chan DK, Chang KW. GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope 2014;124:E34ŌĆō53.


14. Honda K, Griffith AJ. Genetic architecture and phenotypic landscape of SLC26A4-related hearing loss. Hum Genet 2022;141:455ŌĆō64.



15. Rah YC, Kim AR, Koo JW, Lee JH, Oh SH, Choi BY. Audiologic presentation of enlargement of the vestibular aqueduct according to the SLC26A4 genotypes. Laryngoscope 2015;125:E216ŌĆō22.

16. Wu CC, Lu YC, Chen PJ, Yeh PL, Su YN, Hwu WL, et al. Phenotypic analyses and mutation screening of the SLC26A4 and FOXI1 genes in 101 Taiwanese families with bilateral nonsyndromic enlarged vestibular aqueduct (DFNB4) or Pendred syndrome. Audiol Neurootol 2010;15:57ŌĆō66.

17. Chen PY, Lin YH, Liu TC, Lin YH, Tseng LH, Yang TH, et al. Prediction model for audiological outcomes in patients with GJB2 mutations. Ear Hear 2020;41:143ŌĆō9.


18. Sakata A, Kashio A, Koyama M, Urata S, Koyama H, Yamasoba T. Hearing and hearing loss progression in patients with GJB2 gene mutations: a long-term follow-up. Int J Mol Sci 2023;24:16763



19. Wu CC, Lee YC, Chen PJ, Hsu CJ. Predominance of genetic diagnosis and imaging results as predictors in determining the speech perception performance outcome after cochlear implantation in children. Arch Pediatr Adolesc Med 2008;162:269ŌĆō76.


20. Wu CC, Liu TC, Wang SH, Hsu CJ, Wu CM. Genetic characteristics in children with cochlear implants and the corresponding auditory performance. Laryngoscope 2011;121:1287ŌĆō93.


21. Wu CM, Ko HC, Tsou YT, Lin YH, Lin JL, Chen CK, et al. Long-term cochlear implant outcomes in children with GJB2 and SLC26A4 mutations. PLoS One 2015;10:e0138575.

22. Nishio SY, Usami SI. Outcomes of cochlear implantation for the patients with specific genetic etiologies: a systematic literature review. Acta Otolaryngol 2017;137:730ŌĆō42.


23. Wu CC, Lin YH, Liu TC, Lin KN, Yang WS, Hsu CJ, et al. Identifying children with poor cochlear implantation outcomes using massively parallel sequencing. Medicine (Baltimore) 2015;94:e1073.

24. Eppsteiner RW, Shearer AE, Hildebrand MS, Deluca AP, Ji H, Dunn CC, et al. Prediction of cochlear implant performance by genetic mutation: the spiral ganglion hypothesis. Hear Res 2012;292:51ŌĆō8.



25. Hood LJ. Auditory neuropathy/auditory synaptopathy. Otolaryngol Clin North Am 2021;54:1093ŌĆō100.


26. Moser T, Starr A. Auditory neuropathy--neural and synaptic mechanisms. Nat Rev Neurol 2016;12:135ŌĆō49.



27. Lin PH, Hsu CJ, Lin YH, Lin YH, Yang SY, Yang TH, et al. An integrative approach for pediatric auditory neuropathy spectrum disorders: revisiting etiologies and exploring the prognostic utility of auditory steady-state response. Sci Rep 2020;10:9816




28. Wu CC, Hsu CJ, Huang FL, Lin YH, Lin YH, Liu TC, et al. Timing of cochlear implantation in auditory neuropathy patients with OTOF mutations: our experience with 10 patients. Clin Otolaryngol 2018;43:352ŌĆō7.

29. Lin PH, Wu HP, Wu CM, Chiang YT, Hsu JS, Tsai CY, et al. Cochlear implantation outcomes in patients with auditory neuropathy spectrum disorder of genetic and non-genetic etiologies: a multicenter study. Biomedicines 2022;10:1523



30. Hunter JB, Yancey KL, Lee KH. Pediatric single-sided deafness. Otolaryngol Clin North Am 2022;55:1139ŌĆō49.


31. Dhanasingh A, Hochmair I. CI in single-sided deafness. Acta Otolaryngol 2021;141(sup1):82ŌĆō105.

32. Lin PH, Hsu CJ, Lin YH, Lin YH, Lee HY, Wu CC, et al. Etiologic and audiologic characteristics of patients with pediatric-onset unilateral and asymmetric sensorineural hearing loss. JAMA Otolaryngol Head Neck Surg 2017;143:912ŌĆō9.

33. Lee CY, Lin PH, Chiang YT, Tsai CY, Yang SY, Chen YM, et al. Genetic underpinnings and audiological characteristics in children with unilateral sensorineural hearing loss. Otolaryngol Head Neck Surg 2023;169:1299ŌĆō308.


35. Chang MY, Lee C, Han JH, Kim MY, Park HR, Kim N, et al. Expansion of phenotypic spectrum of MYO15A pathogenic variants to include postlingual onset of progressive partial deafness. BMC Med Genet 2018;19:29




36. Chen PY, Tsai CY, Wu JL, Li YL, Wu CM, Chen KC, et al. Hearing features and cochlear implantation outcomes in patients with pathogenic MYO15A variants: a multicenter observational study. Ear Hear 2022;43:1198ŌĆō207.


-
METRICS

-
- 0 Crossref
- 0 Scopus
- 1,278 View
- 16 Download
-
Clinical and Social Outcomes of Cochlear Implantation in Older Prelinguals2023 April;27(2)
Clinical Implication of Inner Ear Anomaly in Cochlear Implantation2006 ;10(1)
The Perception and Production of the Final Question Intonations for the Cochlear Implantee2003 ;7(2)






